Rhizosphere and Proteases
Proteases in plant root exsudates
Roots are not only responsible for nutrient uptake from the soil and anchoring the plant in the substrate but also interact with beneficial as well as harmful microorganisms. Such biotic interactions in the rhizosphere are mediated through plant root exudates and are further influenced by abiotic environmental factors like availability of nutrients.
Plant exudates are usually secreted by vesicle-mediated exocytosis or released from root border cells. Additionally, they might be products of proteolytic events at the root plasma membrane. We identified potential natural proteolytic substrates at the plasma membrane in barley roots. Interestingly, proteases themselves are proteolytic targets as well, which were shown to be still active after release from the plasma membrane.
With specifically designed rhizoboxes, we study protease activity in the rhizosphere of tomato plants by non-invasive soil zymography. The influence of inorganic nitrogen on the distribution of specific proteases along the root system is the focus of our interest.
We identified TREXS, a SDD1-like subtilase in tobacco root exudate that shows high activity with gelatine as substrate (Wendlandt et al., 2015). Subtilases are proteolytic enzymes with key roles in plant development and signal transduction. The physiological role of TREXS remains cryptic but two co-purified proteins provide an informative basis for the identification of in vivo substrates of the subtilase.
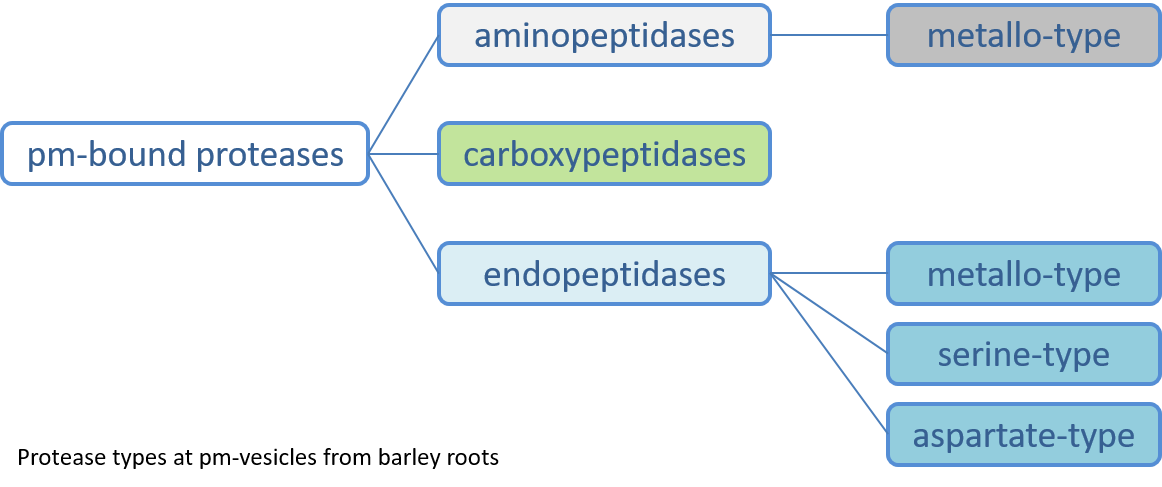
Proteases at the plasma membrane of root cells
Membrane-bound proteases in plants have been located at the inner membranes of plastids and mitochondria as well as at the plasma membrane.Proteolytic processing of proteins at the plasma membrane may create signaling molecules, activate or degrade certain proteins and, thereby, are essential for stress responses and developmental events. In tobacco roots, chromatographically separated plasma membrane-associated proteases displayed different activity patterns dependent on plant nitrogen nutrition (Eick and Stöhr, 2009). By combining detergent treatment, chromatographic and electrophoretic techniques, we identified two types of plasma membrane-bound metallo-aminopeptidases. Gene expression analysis and specific mutants are studied to clarify the function of these exopeptidases and to gain deeper insight into their location.